Every year, influenza – or flu – is responsible for three to five million severe cases and 250,000 to 500,000 deaths worldwide. In other words, the "seasonal" influenza that causes annual outbreaks is far from being a "trivial" infection, and it is by no means harmless. "I didn't realize that flu could knock you out so completely," recalls a former flu sufferer, who was "unable to get out of bed" for six days. The expression "struck down by flu" vividly illustrates the intensity of the symptoms that typically characterize the infection – sudden onset of high fever, muscle pain, headache, generally feeling unwell, dry cough, sore throat and runny nose.
Seasonal influenza affects 3 to 8% of the French population each year, claiming approximately 10,000 to 15,000 lives and worsening the health of already vulnerable populations. More than 18,300 deaths (90% of them in people aged over 65) were reported 2014-2015 winter during the seasonal outbreak, which lasted for nine weeks from mid-January to mid-March, 2015. Some 2.9 million people saw their physician because of flu-like symptoms and 30,000 went to the emergency department suffering from influenza. Over and above the health impact of seasonal flu, it also has major economic consequences, resulting in a loss of between 2 and 12 million working days in France depending on the severity of the outbreak.
A wide variety of viruses
To tackle these seasonal outbreaks, detailed surveillance mechanisms have been introduced at national and international level, making influenza viruses the most closely monitored viruses on the planet (see Interview below). But the task is a difficult one, since influenza is caused by several different viruses. There are three types of influenza virus in humans, A, B and C. Types A and B are responsible for annual outbreaks. Type A viruses are divided into subtypes based on the nature of their surface proteins: hemagglutinin (H1 to H18) and neuraminidase (N1 to N11).

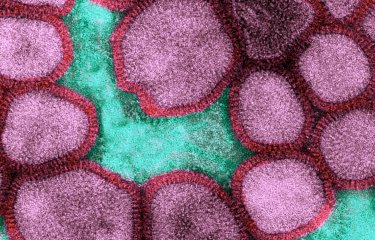
Pandemic 2009 influenza A virus (H1N1) produced in a human muscle cell culture. Colorized electron microscopy.
© Institut Pasteur/Marie-Christine Prévost, Ultrastructural Microscopy Platfrom - Marion Desdouits and Pierre-Emmanuel Ceccaldi, Oncogenic Virus Epidemiology and Pathophysiology Unit, Sylvie Van der Werf et Nadia Naffakh, Molecular Genetics of RNA Viruses Unit. Colorization by Jean-Marc Panaud.
The most widespread virus in France during the last winter outbreak in 2016-2017 was an A virus (H3N2). To complicate things even more, there are variants within each subtype, since influenza viruses are permanently evolving and new strains are constantly emerging (this explains why the vaccine changes each year). The nomenclature for influenza viruses is based on their geographical origin and year of isolation.
For example, the strain of virus A(H3N2) used in the composition of the vaccine for the 2017-2018 season is named A/Hong Kong/4801/2014 (H3N2). Each year, the World Health Organization (WHO) recommends which strains should be used to compose the vaccine (three in France and four in other countries) on the basis of the information it receives from the different countries on the viruses circulating there. This information is used to identify the most common strains in circulation and any new variants that the population needs to be protected against.

The global surveillance network for influenza viruses is undoubtedly the oldest and best structured virological surveillance network.
The Institut Pasteur plays a key role in monitoring influenza in France. How does this surveillance work in practice?
We coordinate the National Reference Center for Influenza. In mainland France, we have 1,300 family physicians in the "Sentinelles" network [a computerized information system for communicable diseases] who monitor patients with feverish flu-like symptoms and take samples. Around 2,000 samples are sent on average each year to the Institut Pasteur for northern France and to a CNR-associated laboratory in Lyon for southern France. We use these samples to search for influenza viruses and also systematically for other major respiratory tract viruses, such as respiratory syncytial virus (RSV).
We characterize the influenza viruses and determine their genome sequences to see how they are evolving and whether they still match the vaccine. We also receive data from the RENAL hospital laboratory network for an extensive list of respiratory agents (numbers of diagnoses and positive detections, etc.). This gives us an overview of what is circulating in hospitals and the impact it is having. Each week, we publish a bulletin with reports on the current situation concerning influenza and other respiratory tract infections. Hospitals also send us influenza virus strains from severe cases or treatment failures so that we can analyze their sensitivity to antiviral drugs. This national virological surveillance is supervised by Santé publique France, which also monitors statistics such as the death rate from influenza and numbers of visits to hospital emergency departments or calls to SOS Médecins.
How do these national measures fit in with global surveillance efforts?
It's like a set of Russian dolls! Our strategy is part of the EU network, led by the European Center for Disease Prevention and Control (ECDC) in Sweden. The ECDC is itself part of the "EuroFlu" network for the broader geographical European region, coordinated by WHO/Europe. This in turn is part of the influenza network of the World Health Organization. Every country sends one of the five global WHO Centers (in London, Atlanta, Melbourne, Tokyo and Beijing) a selection of the seasonal viruses circulating within its borders, so that they can compare the viruses in the different countries. We work with the London Center, and we generally send two batches each year. Ultimately all the information feeds back into WHO, and comparative analyses are performed to help determine the composition of the next year's vaccine. This is decided in February for the vaccine released in October for the northern hemisphere, and in September for the vaccine launched in April in the southern hemisphere. This structure also makes it immediately obvious if any new viral variants emerge or spread, so that the alert can be sounded quickly. As soon as we observe a virus that is out of the ordinary, we immediately send it to London.
Can it be said that influenza viruses are the most closely monitored viruses on the planet?
The global surveillance network for influenza viruses is undoubtedly the oldest and best structured virological surveillance network. It is also used to monitor other emerging respiratory tract viruses. It was a logical decision to make use of the influenza network in our efforts to tackle the SARS outbreak in 2003. In 2013, the Institut Pasteur confirmed two cases of Middle East respiratory syndrome coronavirus (MERS-CoV) in northern France. There have been no new cases since then, but we regularly analyze "possible" cases. Getting back to influenza, we also analyze suspected human cases of avian influenza, H5N1 or H7N9, and of a type of swine flu in the United States, but alerts are fortunately rare.
In the event of a health crisis, the Influenza CNR is supported by the Institut Pasteur's Laboratory for Urgent Response to Biological Threats (CIBU), directed by Jean-Claude Manuguerra. During the 2009 pandemic, the CIBU worked non-stop, seven days a week, from late April to late August.



This structure makes it immediately obvious if any new viral variants emerge or spread, so that the alert can be sounded quickly. As soon as we observe a virus that is out of the ordinary, we immediately send it to the WHO Center.
Vincent EnoufResearcher in the Molecular Genetics of RNA Viruses Unit and Deputy Director of the French National Reference Center for Influenza at the Institut Pasteur.© Institut Pasteur - François Gardy photo
Falling vaccine coverage
The flu vaccine, which therefore changes each year, is the best means of protection against influenza. It is not only advisable but strongly recommended and refunded for those aged over 65 or belonging to one of the risk groups identified by the French High Council for Public Health (HCSP), which include pregnant women, obese individuals and adults and children with certain chronic cardiovascular or respiratory conditions. Influenza is a severe disease for these populations, and the health authorities regret the fall in vaccination rates: "The drop in vaccine coverage is worrying; the rate fell to 46% in 2016-2017, from 47% in 2015-2016 [and as high as 52% in 2013-2014]," emphasized Santé publique France (SpF) in its report on the latest seasonal outbreak. Some vaccination critics claim that the vaccine is not effective. But even if the match between circulating and vaccine strains was not perfect in 2016-2017, most circulating influenza viruses were covered by the vaccine, as pointed out by SpF in March 2017. SpF also noted that preventing influenza and its complications involves both "vaccination and also the adoption of preventive measures (such as keeping away from people at risk and stepping up basic hygiene measures), as well as the use of antiviral drugs for those at risk of complications."

Good health habits can also help prevent flu
In addition to vaccination, basic hygiene measures can help stop the spread of influenza. While flu mainly spreads through the air via tiny particles of saliva and especially droplets released when coughing and sneezing, influenza viruses also have a degree of resistance to the external environment, meaning that people can sometimes catch flu by touching an object with the flu virus on it, such as a door handle. It is therefore highly recommended that you protect yourself by:
- washing your hands with soap and water after coming into contact with flu sufferers;
- avoiding getting too close to anyone with flu;
- cleaning any objects used regularly by infected people.
It is also important to protect others when you are suffering from flu, by covering your mouth when you cough and your nose when you sneeze, blowing your nose with disposable paper tissues and throwing them away in a bin with a lid, and washing your hands after all of the above.

Influenza A virus with spicules bristling the surface of virions. Transmission electron microscopy. Colorized image. © Institut Pasteur / Charles Dauguet
Pandemic threat
These preventive measures should be used both to curb the spread of seasonal flu and to reduce the risk of a pandemic. There are constant fears that a new virus will emerge that is capable of spreading rapidly across the world. The 20th century witnessed three major flu epidemics: "Spanish" flu (1918-1920, 40 million deaths (see inset below)), Asian flu (1957-1958, 1 to 1.5 million deaths) and Hong Kong flu (1968-1969, 1 million deaths). And the first pandemic of the 21st century, caused by influenza A(H1N1)pdm09 virus (2009-2010, 400,000 deaths) – which was fortunately less serious than expected (see inset below) – will probably not be the last.
A look back at the Spanish flu pandemic
The sudden demise of the Cyrano de Bergerac author illustrates the violence of the "Spanish" flu, which also claimed the lives of French poet Guillaume Apollinaire, Austrian painter Egon Schiele and 40 million others worldwide (with more than 400,000 in France alone).
This influenza outbreak was more deadly than the First World War, spreading over the entire planet in the space of a few months and affecting more than a third of the global population between 1918 and 1919. It was referred to as "Spanish flu" because Spain, a neutral country and therefore not bound by military secrecy in wartime, was the first country to mention the epidemic publicly. But it is actually thought that this pandemic, the deadliest in history over such a short space of time, had its origins in China, before spreading to the United States on a US battalion returning from Guangzhou to a base in Boston. The first deaths were reported in US military camps in February 1918. The virus is believed to have mutated in the United States, becoming more virulent and deadly, and then to have been introduced in Europe that spring with the arrival of American troops. It soon spread throughout the rest of the world via trade between the European powers and their colonies.
The 1918 A(H1N1) virus (a remote ancestor of the virus that caused the 2009 pandemic) was characterized in the 1990s by Jeffery Taubenberger's team, which analyzed tissue samples preserved in paraffin from collections belonging to the US military health service, taken from two soldiers who died in 1918. The same team then revealed the whole genome sequence of the virus in 2005, using viral RNA isolated from the lungs of an Inuit woman, another victim of the 1918 pandemic, who was exhumed from the Alaskan permafrost (deep layers of soil or rock that remain frozen all year round) by Swedish pathologist Johan Hultin. Although we now understand a lot more about the pandemic, we still do not know why the 1918 virus was so virulent. Elucidating the mystery would help us to evaluate the risks associated with other strains of influenza. The scientific investigation goes on...
"During the final days of November 1918, Edmond Rostand left his house in Cambo-les-Bains and went to Paris to join in the festivities and soak up the celebratory atmosphere following the end of the First World War. One evening he went to the Sarah Bernhardt Theater to watch a rehearsal of L'Aiglon starring the great actress. He caught a chill in the wings and went home shivering and suffering from chest pains. The next day, November 30, his temperature reached 41°C. Two days later he was dead."
In La grippe “espagnole” en France en 1918-1919 (The "Spanish" Influenza in France in 1918-1919) by Jean Guénel; Histoire des sciences médicales.

Why did the 2009 H1N1 virus not have the impact that people had initially feared?
"This was largely because, very fortunately, there was a higher level of pre-existing immunity to this virus than we could have expected," explains Prof. Sylvie van der Werf, Head of the Molecular Genetics of RNA Viruses Unit at the Institut Pasteur. "Although the population was naive to this new virus, there was some immunity, especially in elderly people, since the A(H1N1)pdm2009 virus contained elements derived from the 1918 A(H1N1) virus, which remained in circulation until 1957. Other viruses derived from the 1918 virus also circulated among the population for a lengthy period. Many people had been exposed, to a greater or lesser extent, to other H1N1 viruses which induced a degree of immunity that provided some protection against the 2009 H1N1 virus. No one had measured the scale of this residual immunity. But the 2009-2010 pandemic was by no means a minor outbreak, as is sometimes claimed. Yes, the impact was less severe than had been feared, but it was still a virus of pandemic proportions, which resulted in serious and fatal cases in young people to an extent that we had never witnessed before with seasonal outbreaks." The A(H1N1)pdm2009 virus is still in circulation, but it has become seasonal in nature, and one of its strains is now used in the composition of the annual vaccine.
Animal reservoirs

All these pandemics were caused by type A viruses. These infect humans and also several animal species, such as poultry, pigs, horses and a number of other mammals. Waterfowl such as wild ducks serve as natural reservoirs for nearly all known subtypes of influenza A, and they do not display any symptoms. They can transmit these viruses to domestic poultry and also to pigs and horses – which, when infected, suffer from respiratory symptoms like humans. During their time in different animals – especially in pigs, which can serve as a crucible –, the viruses can mingle and exchange genetic material (through a process known as reassortment).
The genome of the A(H1N1) virus responsible for the 2009 epidemic, for example, was an entirely new combination of genes from bird, human and swine viruses.

Persistence of influenza viruses in the environment
How long can influenza viruses persist on a smooth surface? "Up to 66 days at 4°C, and 3 days at 35°C," says Jean-Claude Manuguerra, Head of the Environment and Infectious Risks Unit at the Institut Pasteur. His team has tested the behavior of these viruses on watch glasses and is currently studying the persistence of avian viruses in water. "We carry out laboratory simulations, varying the water temperature and salinity to mimic natural conditions in the seas and oceans, up to the extreme conditions in the Dead Sea, for example," says the scientist. "We have shown that viruses in waterfowl can persist for more than a thousand days in fresh water at 4°C, like the water at the bottom of the Siberian lakes. Our findings suggest that the environment can serve as a "reservoir" for these viruses: they can potentially persist in lakes until the next migration of birds the following year." By comparing viruses with different levels of resistance, the team set out to find out what confers persistence on these influenza viruses at molecular level. Their research highlighted the importance of the two glycoproteins at the viral surface, hemagglutinin (a key protein that enables the virus to enter cells) and neuraminidase, which alone are capable of determining the stability of influenza viruses. The frequent naturally occurring mutations in hemagglutinin therefore affect the stability of influenza viruses in the environment. We now need to shed light on how they influence the spread of the virus.
Avian and swine influenza
Some viruses already found in animals can also be transmitted directly to humans. Since 2003, the avian influenza A(H5N1) virus, responsible for a vast "epizootic" which killed millions of birds, has infected 860 people and claimed 454 lives, mainly in Egypt, Indonesia and Vietnam. This virus, highly pathogenic in birds, has the potential to mutate and develop the ability to spread easily from one individual to another. The same is true of other closely monitored viruses, such as avian influenza A(H7N9), which since 2013 has affected several people in China (1,564 cases and 612 deaths), and a swine flu virus in the United States. But the next pandemic may come from somewhere completely unexpected.