The structure of DNA and chromosomes is a major focus of research, but still holds a number of mysteries. Scientists from the Institut Pasteur and the CNRS have developed a new method for quantifying chromatin loops and observing chromosomes at high resolution.
In our cells, DNA is coiled up many times, enabling it to fit into the cell nucleus, which is approximately 200,000 times smaller than the length of the uncoiled DNA. DNA is initially coiled around proteins known as histones, to form a structure called the nucleosome. Nucleosomes in turn form a structure known as chromatin fiber. We have known for some years that chromatin fiber forms loops that are maintained by a molecule complex called cohesin. These chromatin loops help structure chromosomes and play an important role in basic genome functions such as gene expression.
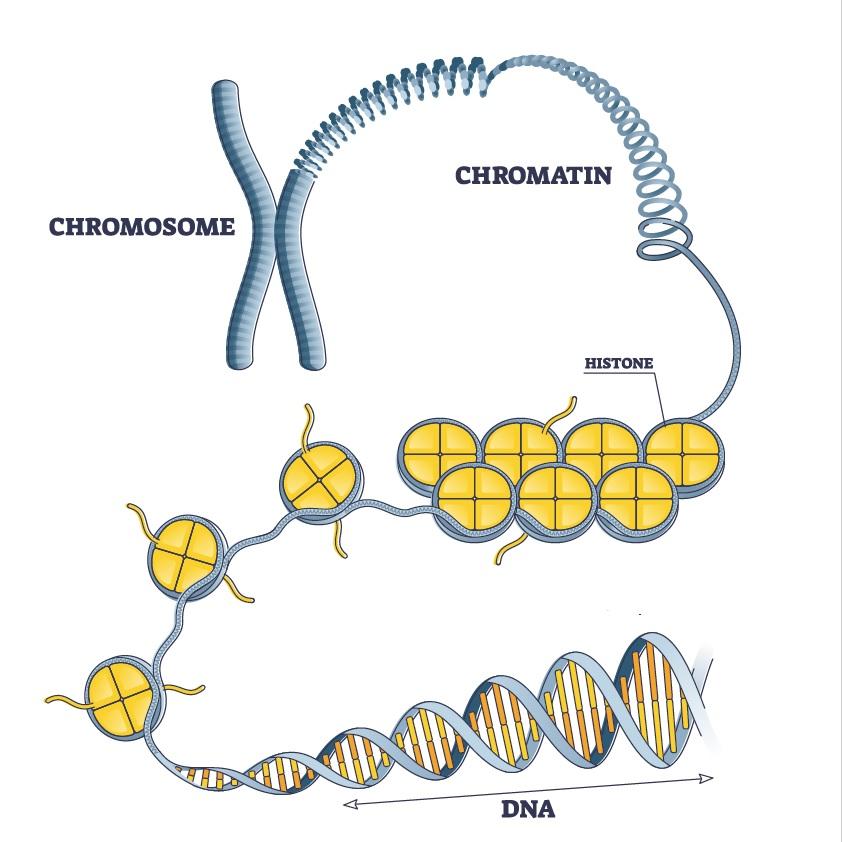
Highly simplified diagram of DNA compaction in the chromosome. Here the chromosome is symbolized in the form of an X (metaphase). For the article published on May 11, 2021, the Institut Pasteur's scientists studied DNA in another phase of the cell cycle (interphase), where chromosomes have a different form (one that could be described as "spaghetti").
A new approach to observing chromatin
The organization of chromatin in the cell nucleus is currently mainly studied with the Hi-C technique, which uses sequencing to quantify contacts between different DNA regions. "Hi-C generally provides averages for large populations of cells; it cannot be used to characterize the presence or number of loops for individual cells. We developed an approach combining imaging with polymer simulations to describe the chromatin structure in individual human cells," explains Christophe Zimmer, Head of the Imaging and Modeling laboratory at the Institut Pasteur.
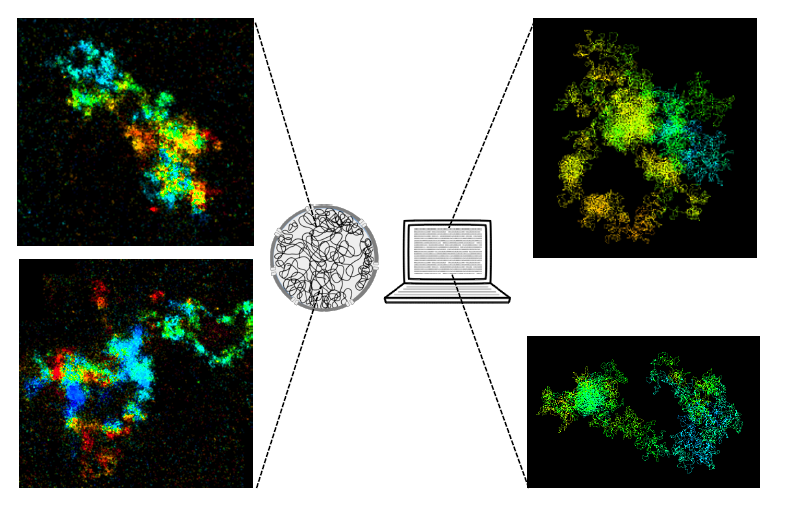
Imaging and modeling human chromosomes. On the left, two images of chromosomes observed using high-resolution 3D microscopy. On the right, two images of chromosomes generated using a computer simulation.
The majority of human DNA enclosed in loops maintained by cohesin
This new approach can be used to visualize chromosomes or chromosome regions at high resolution, in isolation from each other, in intact nuclei without DNA denaturation. The scientists used this method to analyze the influence of cohesin on the chromatin structures. Their analyses suggest the simultaneous presence of tens or hundreds of thousands of loops in the genome of individual cells, indicating that the vast majority of human DNA is enclosed in loops maintained by cohesin.
This study provides new information about basic genome organization structures, as well as a method to visualize and model the detailed structure of chromatin.
Source
Super-resolution visualization and modeling of human chromosomal regions reveals cohesin-dependent loop structures, Genome Biology, 11 mai 2021
Xian Hao1,2†, Jyotsana J. Parmar1,3†, Benoît Lelandais1†, Andrey Aristov1, Wei Ouyang1,4, Christian Weber1 and Christophe Zimmer1*
†Xian Hao, Jyotsana J. Parmar and Benoît Lelandais contributed equally to this work.
1Institut Pasteur, Imaging and Modeling Unit, UMR 3691, CNRS, Paris, France
2School of Public Health, Nanchang University, Nanchang 330006, China.
3Simons Center for the Study of Living Machines, National Center for Biological Sciences (TIFR), Bangalore, Karnataka 560065, India.
4Université Paris Diderot, PSL University, F-75005 Paris, France.