Calmodulin is a protein produced naturally by the body whose activity is modulated by the calcium in our cells. Scientists at the Institut Pasteur have elucidated the mechanisms of interaction between calmodulin and calmidazolium. The latter is able to effectively reduce the biological effects of calmodulin, but it is also toxic for cells. This study paves the way for the development of new, more selective calmodulin inhibitors with fewer adverse effects.
Calmodulin is a protein found in all living organisms, or at least those whose cells have a nucleus. "Calmodulin" literally means "calcium-modulated protein," and it is a key element in cellular metabolism. This means that it is involved in a variety of biological functions, including cell architecture and movement, cell division, and intercellular communication. It is also involved in conditions such as cardiac arrhythmias and the development of Alzheimer's disease. Calmodulin is hijacked by bacterial toxins in cell intoxication processes (see Mechanism of activation of a major toxin involved in whooping cough – Institut Pasteur news article). Acting on calmodulin is therefore a key focus for basic research and health care.
Calcium, calmodulin, calmidazolium: poorly understood interactions
We already know that by binding to calmodulin, calcium changes the molecular conformation, turning calmodulin into a protein "switch" that regulates a whole series of other proteins. Despite its important role in cell function, the structure and dynamics of calmodulin are still a subject of much discussion and research.
One of the most extensively researched calmodulin antagonists is calmidazolium, discovered in 1981. It can easily bind to calmodulin to inhibit its physiological effects. "Calmidazolium, like many other calmodulin antagonists, is unfortunately known for its cytotoxicity and its interactions with other cell targets," explains Alexandre Chenal, Head of the BiophysiCyaA group. The scientist previously worked on the links between calmodulin and the CyaA toxin, a major virulence factor of the whooping cough agent, in the Biochemistry of Macromolecular Interactions Unit led by Daniel Ladant at the Institut Pasteur. Until now, no one had successfully resolved the structure of the calmodulin-calmidazolium complex. The mechanisms underpinning this affinity were also poorly defined, with data varying from one publication to the next. Alexandre Chenal's team used various technological approaches to try to answer these questions.
State-of-the-art technologies to measure the effects of calmidazolium
The scientists used a technique known as SRCD at the SOLEIL synchrotron facility. "We demonstrated that calmidazolium does not alter local folding of calmodulin," explains Corentin Léger, a post-doctoral fellow in the BiophysiCyaA research group and first author of the paper.
Using a quantification method known as ITC, with the Institut Pasteur's Molecular Biophysics Platform, the scientists also determined the affinity of the calmodulin-calmidazolium interaction and showed that binding of a single calmidazolium molecule is sufficient to inhibit calmodulin. The next task was to determine whether the interaction between calmodulin and its inhibitor changes the spatial conformation of calmodulin.
To this end, the scientists employed various other analytical techniques:
- SEC-SAXS (size-exclusion chromatography small-angle X-ray scattering) to study the spatial conformation of the calmodulin-calmidazolium complex – a technique performed at the SOLEIL synchrotron;
- HDX-MS (hydrogen deuterium exchange mass spectrometry) and NMR (nuclear magnetic resonance) to better characterize the calcium-calmodulin complex – two techniques carried out by one of the Institut Pasteur's technological platforms.
The scientists revealed that in the presence of calmidazolium, calmodulin – initially extended and flexible – took on a compact, rigid conformation. Calmidazolium acts as a "glue" in the molecule, maintaining calmodulin in a closed conformation (see figure below) and rendering it unavailable for interacting with its physiological partners.
Using an X-ray-based crystallography technique (with the Crystallography Platform and the SOLEIL synchrotron), the scientists successfully determined the atomic structure of the complex formed by calmodulin and calmidazolium. The NMR and HDX-MS methods also demonstrated that binding to calmidazolium reduces the dynamics of calmodulin and sets it in a compact, closed conformation (see figure).
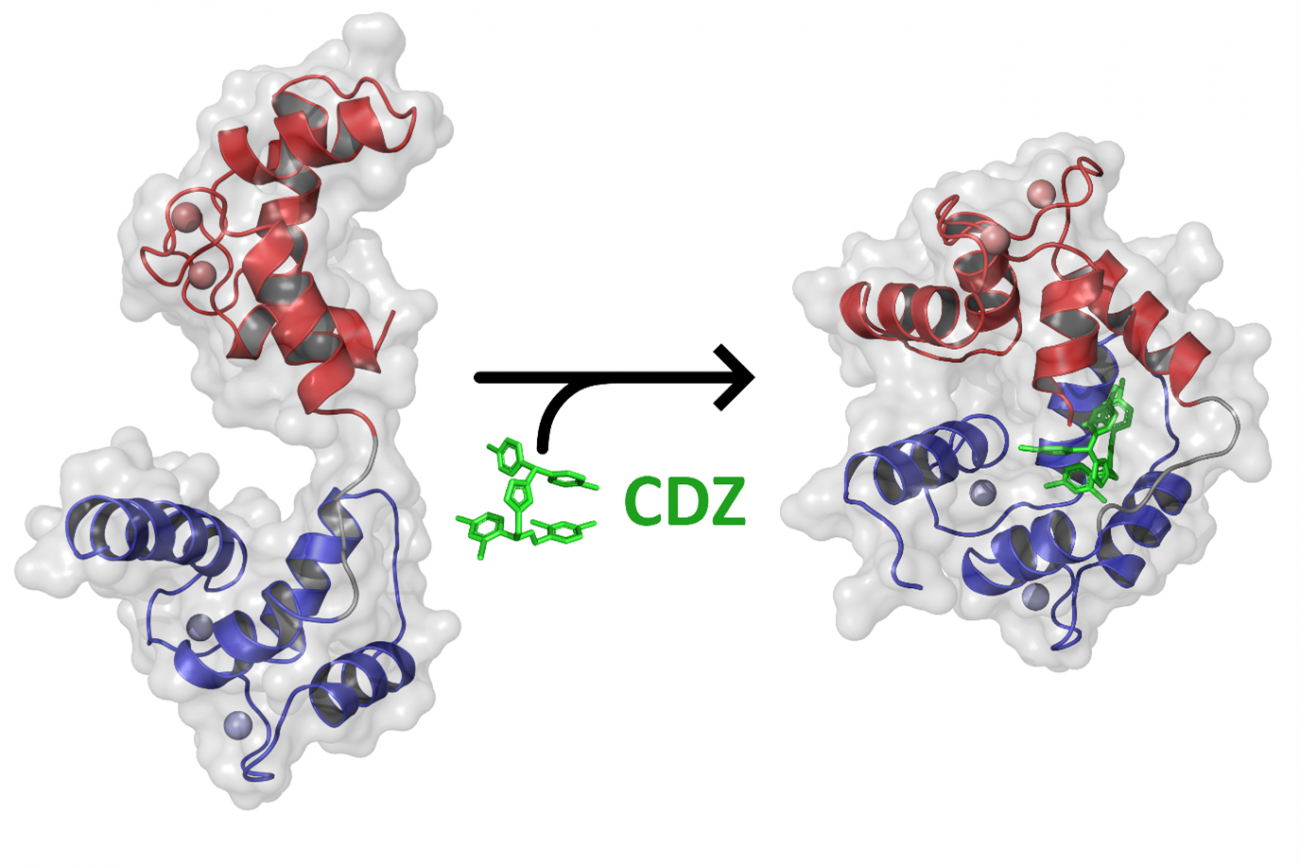
Addition of an excess of calmidazolium does not change the overall structure of the calmodulin-calmidazolium complex. A single molecule of calmidazolium is sufficient to change the conformation of calmodulin and inhibit it.
As explained above, calmidazolium does not only bind to calmodulin; it also interacts with other cellular components, giving rise to adverse effects. This research should enable chemists and bioinformaticians to develop new, more specific calmodulin inhibitors that are less toxic for cells. "These new molecules will be useful in fundamental research to study calmodulin, and over the longer term they could potentially represent candidates to treat diseases involving calmodulin," concludes Alexandre Chenal.
The study also illustrates the synergies between the Institut Pasteur's research teams and its technological platforms, especially the Molecular Biophysics, Crystallography and Biological NMR Platforms, as well as the DISCO, SWING and PROXIMA beamlines at the SOLEIL synchrotron in Saint-Aubin, France.
Source
Dynamics and structural changes of calmodulin upon interaction with its potent antagonist Calmidazolium, BMC Biology, August 09, 2022
Corentin Léger1, Irène Pitard2, Mirko Sadi1,3, Nicolas Carvalho1,3, Sébastien Brier2, Ariel Mechaly4, Dorothée Raoux-Barbot1, Maryline Davi1, Sylviane Hoos5, Patrick Weber4, Patrice Vachette6, Dominique Durand6, Ahmed Haouz4, J. Iñaki Guijarro2, Daniel Ladant1, Alexandre Chenal1
1: Biochemistry of Macromolecular Interactions Unit, Department of Structural Biology and Chemistry, Institut Pasteur, Université Paris Cité, CNRS UMR3528, 75015 Paris, France
2: Biological NMR and HDX-MS Technological Platform, Institut Pasteur, Université Paris Cité, CNRS UMR3528, 75015 Paris, France
3: Université Paris Cité, Sorbonne University, Paris, France
4: Crystallography-C2RT Platform, Institut Pasteur, Université Paris Cité, CNRS UMR3528, Paris, France
5: Molecular Biophysics Platform, Institut Pasteur, Université Paris Cité, CNRS UMR3528, Paris, France
6: Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France