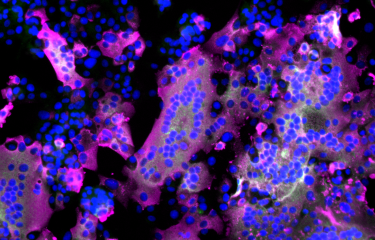
The COVID-19 pandemic, which has afflicted the world for more than two years, is caused by SARS-CoV-2. The genome of this respiratory virus is composed of a single strand of RNA, a molecule similar to DNA that enables the virus to replicate in the body. Scientists from the Institut Pasteur, Inserm and the Paris Public Hospital Network (AP-HP) have developed a new method, CoronaFISH, to observe the virus and detect its presence.
Since the start of the pandemic, the ability to study the biology of the virus in cells and detect its presence in infected organs or patients has been crucial. This requires microscopy visualization tools. "Several techniques currently exist, but most have limitations – they may be indirect, expensive or unreliable. For example, we can use antibodies to reveal viral proteins, but obtaining these antibodies is a long and expensive process, their sensitivity is compromised by variants, and the presence of viral proteins in a cell does not necessarily imply that the virus is actively replicating there," explains Christophe Zimmer, Head of the Imaging and Modeling laboratory at the Institut Pasteur.
To visualize the viral genome itself, scientists from the Institut Pasteur, Inserm and the AP-HP have developed a new method. The aim of this method, known as CoronaFISH, is to label viral RNA directly based on fluorescence in situ hybridization (FISH), a technique that can be used to identify a specific part (or sequence) of RNA by binding to its components, nucleotides, via complementarity. Each RNA sequence has a complementary sequence which naturally binds to the RNA molecule. "This method uses a series of probes made up of DNA fragments linked to fluorescent molecules that are complementary to the viral RNA sequence", says Florian Mueller, a scientist in the same laboratory and one of the lead authors of the article. "If the virus is present, the oligonucleotides bind to the viral RNA, which then becomes visible by fluorescence under a microscope", he continues.
The scientists validated CoronaFISH in African green monkey cells, which are frequently used to study SARS-CoV-2, and in human cells. "Our technique was capable of distinguishing cell populations with different viral loads, even if only some cells were infected. Unlike methods that analyze cell populations more generally, CoronaFISH can be used to perform a spatial and temporal analysis of SARS-CoV-2 replication and propagation," comments Giovanna Barba-Spaeth from the Structural Virology Unit, one of the senior authors of the paper.
They also demonstrated that CoronaFISH can detect the virus in the lung tissue of a person with COVID-19, as well as in nasal swabs used for PCR tests. Finally, the team modified CoronaFISH so that it could be used to observe SARS-CoV-2 via electron microscopy, making it possible to study the virus in the context of the cell ultrastructure (the detailed structure of elements that compose the cell).
This method expands the toolbox available to study SARS-CoV-2, enabling viral RNA to be visualized in detail using optical or electron microscopy, in cells or tissues. It will therefore help improve our understanding of which organs can be affected by the virus.
Once the method has been optimized and validated by further research, it could even serve as the basis for a diagnostic test. A test based on CoronaFISH would have various advantages compared with PCR: for example, it would not require RNA extraction, and given the large number of probes used, its sensitivity should not be hindered by the emergence of variants.
This research received funding from AXA COVID-19 project COVID-SPREAD.
The Institut Pasteur Center for Translational Science provided support for the project's ethical and regulatory compliance. The project was also registered on the Health Data Hub platform which grants the public access to research projects using personal data that may originally have been collected during healthcare provision for instance.
This study is part of the priority scientific area Emerging infectious diseases of the Institut Pasteur's strategic plan for 2019-2023.
Source:
Sensitive visualization of SARS-CoV-2 RNA with CoronaFISH, Life Science Alliance, January 7, 2022
Elena Rensen1,Stefano Pietropaoli2, Florian Mueller1, Christian Weber1, Sylvie Souquere3, Sina Sommer2, Pierre Isnard4,5, Marion Rabant4,5, Jean-Baptiste Gibier6, Fabiola Terzi4, Etienne Simon-Loriere7,Marie-Anne Rameix-Welti8,9, Gerard Pierron10, Giovanna Barba-Spaeth2, Christophe Zimmer1
1 Institut Pasteur, Université de Paris, CNRS UMR 3691, Imaging and Modeling Unit, Paris, France
2 Institut Pasteur, Université de Paris, CNRS UMR 3569, Unité de Virologie Structurale, Paris, France 3 Gustave Roussy, AMMICA UMS-3655, Villejuif, France
4 Université de Paris, INSERM U1151, CNRS UMR 8253, Institut Necker Enfants Malades, Département “Croissance et Signalisation”, Paris, France
5 Service d’Anatomo-Pathologie, AP-HP, Hôpital Necker Enfants Malades, AP-HP Centre, Paris, France
6 Service d’Anatomo-Pathologie, Centre de Biologie Pathologie, CHU Lille, Lille, France
7 Institut Pasteur, Université de Paris, G5 Evolutionary Genomics of RNA Viruses, Paris, France
8 Université Paris-Saclay, INSERM, Université de Versailles St. Quentin, UMR 1173 (2I), Montigny-le-Bretonneux, France
9 AP-HP, Université Paris Saclay, Hôpital Ambroise Pare, Laboratoire de Microbiologie, Boulogne-Billancourt, France
10 Gustave Roussy, CNRS UMR 9196, Villejuif, France