A historic fight against emerging infectious diseases

Emerging infectious diseases result from contact between microbes and animals, with humans the most parasitized animal species. From the plague in Asia in 1894 to the current COVID-19 crisis, scientists at the Institut Pasteur have long leveraged their expertise and talent to further knowledge creation and benefit humankind.
Emergence, an inevitable and unpredictable fact of life
Between 1918 and 1919, in just a few months a third of the world's population was infected with Spanish flu. While Spain was the first country to report the disease, the outbreak probably started in Asia and subsequently spread to the United States before reaching Europe in a more virulent and lethal form with American soldiers. The risk of pandemics is higher now than in 1918 due to the speed of travel. Viruses can travel the world in a day, compared to a year in the 19th century. While research and public health responses are sometimes slowed by regulations, alert systems have improved and we are now better informed about virus biology. Moreover, laboratory testing capacity has increased following the introduction of the International Health Regulations in 2005.
We also know that the introductory phase (or spillover – see below) is critical for tackling outbreaks. In 2003, rapid identification of the severe acute respiratory syndrome (SARS) virus led to effective containment measures as patients were not infectious prior to symptom onset, and it was thus possible to eradicate the virus. In contrast, COVID-19 spread quickly in 2020 following its introductory phase. WHO thus declared a pandemic, triggering an immediate response from the scientific community. A combination of vaccinations and lockdown measures then helped limit disease severity and spread.
Why are we still faced with such a grave risk? An analysis of key factors in viral emergence since 1940 suggests that changes in land use play a dominant role in 1 in every 4 cases. While climate appears to have less of an impact, except in relation to arboviruses, this situation may well change with global warming.
What are the main causes? By definition, emerging viruses are unpredictable. However, often respiratory and easily transmissible RNA viruses such as influenza and coronaviruses present the greatest risk. In contrast, AIDS was caused by a retrovirus that emerged in the 1980s. The key takeaway is that emergences are an "inevitable fact" in the words of scientist Charles Nicolle. However, let's not be fatalistic as scientific progress is a constant source of hope.
With Jean-Claude Manuguerra, head of the Environment and Infectious Risks Unit, Laboratory for Urgent Response to Biological Threats and Hantaviruses CNR, and co-head of the Emerging Animal Pathogens in Humans OIECC (World Organization for Animal Health Collaborating Center).
What is an emerging infectious disease?

emergence, noun.
Sudden appearance in a series of events or ideas. By extension, the appearance of a new biological system or new properties. Synonyms: occurrence, appearance, surge.
An emergence pre-exists its discovery and only appears to those who encounter it.
In human health, emergence points to a revelation. "An emerging disease denotes the sudden appearance within a human population of a novel pathogen from an animal or environmental reservoir or following the genetic modification of an existing pathogen," explains Caroline Demangel. "It may also denote a known disease that reappears by extending its geographical range or becoming more transmissible or severe. This is referred to as a reemerging disease." Emerging and reemerging infectious diseases are epiphenomena of human existence and our interactions with each other, and with nature. As human societies grow in size and their habitats encroach on natural ecosystems, we create an infinite variety of opportunities for infectious agents to transmit to humans and for new diseases to emerge. The meaning of the term "emergence" implies surprise and a sudden change from a known situation. But how do we prepare for the unknown? That's what scientific research is all about.
With Caroline Demangel, head of the Immunobiology and Therapy Unit and co-head of the Emerging infectious diseases priority area (priority area 1 of the Institut Pasteur 2019-2023 Strategic Plan).
Welcome to the age of emergence
In 1995, Stephen Morse, Professor of Epidemiology at Columbia University put forward his definition of emerging infectious diseases as "infections that have newly appeared in a population or have existed but are rapidly increasing in incidence or geographic range." His theorization came at the end of a century in which optimistic visions of eradicating infectious diseases encouraged by successes including the eradication of smallpox declared by the World Health Organization (WHO) in 1979, coexisted with a rise in the number and scale of emerging or reemerging infectious diseases such as Ebola and AIDS. While there is no doubt that our knowledge of these phenomena and ability to respond have improved, this can nevertheless be described as the "age of emergence" due to the dramatic increase in causative factors. No fewer than 335 emerging diseases were detected between 1940 and 2004 affecting all continents. How has this happened?
 On June 15, 1894, Alexandre Yersin landed in Hong Kong with a microscope and autoclave in the midst of an epidemic that was decimating the population. In his straw hut (see photo), he used a bubo specimen taken from a deceased patient to identify the plague bacillus which was later named Yersinia pestis. For over 130 years, a whole host of Institut Pasteur scientists have studied endemic and epidemic diseases. |
Anthropocene: the fourth upheaval
Emerging infectious diseases are the result of encounters in which humans top all the rankings – 1,400 species of microbes and parasites were listed as potentially affecting Homo sapiens at the start of the 21st century (Cleveland et al., 2001), making it the most parasitized animal species.
The first upheaval occurred around 75,000 BCE, with the Neolithic Age which marked the start of social and economic organization. Hunter-gatherers began to form small communities and settle. Farming, domestication of animal species, interaction with wild fauna, and proximity among individuals created the first conditions conducive to cross-species transmission. With population spread came increasing opportunities for transmission, heralding the emergence of diseases such as whooping cough, tuberculosis and measles.
The second upheaval, which occurred around 1 CE, was driven by urbanization and trade prompted by the rise of the great Roman and Chinese empires. Economic links forged between Europe and Asia precipitated the microbial unification of Eurasia. As caravans traveled the silk roads, people began to mix, enabling the exchange and circulation of goods, ideas and practices, but also of diseases such as plague, smallpox and leprosy.
The third upheaval occurred at the turn of the 16th century driven by major naval conquests and the expansion of colonial empires. Microbial unification took on a transcontinental dimension between the old and new worlds. Colonial conquests followed by triangular trade led to infectious diseases such as smallpox, measles, syphilis and malaria spreading on both sides of the Atlantic.
The boom in trade and globalization that shaped the history of the 20th century was the final step cementing the advent of the fourth upheaval, the Anthropocene, in which human influences are the primary driver of change in ecosystems. A virus can now switch continents overnight on an international flight…
However, while it is possible to interpret the history of humanity through the lens of trade, and therefore understand how its intensification contributed to the emergence of infectious diseases, it is also necessary to examine factors promoting the appearance of pathogens among humans.
How and why do infectious diseases emerge?
While numerous mechanisms are involved in the emergence of infectious diseases, the process can be divided into three main steps.
Stage 1 : spillover
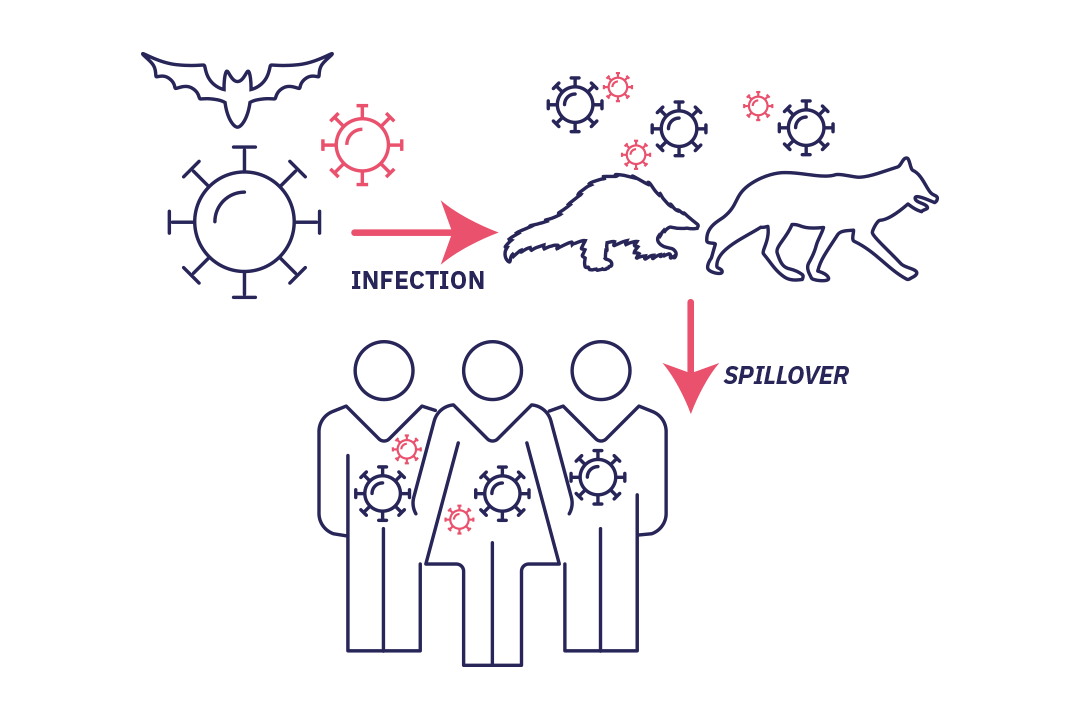
Stade 1 : Spillover without secondary transmission: (accidental) infection of a new host without the virus being able to develop in this new human host.
It starts with the accidental infection of a host by a microorganism. The biological agent is introduced to the host (often an animal) or its secretions (saliva, urine, etc.), feces or the contaminated environment. A human then comes into contact with the animal, either directly or through a second animal (vector). Infection subsequently occurs. This is known as spillover, where a virus "spills over" from its usual habitat and is transmitted to humans. In most cases, the virus is unable to develop in its new human host and the story ends there.
Stage 2 : cross-species transmission
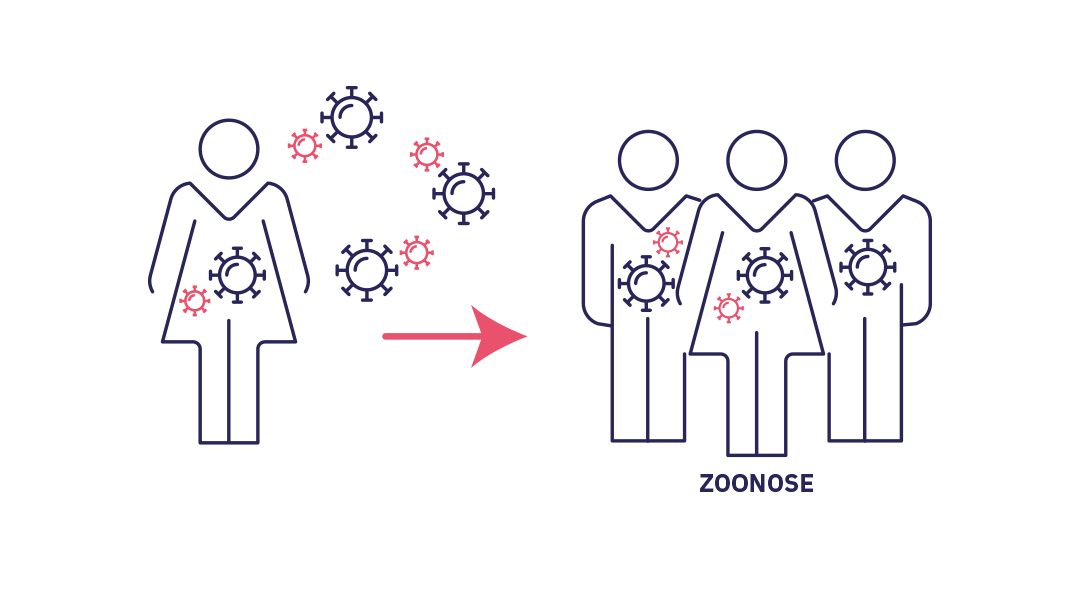
Stade 2 : The pathogen becomes established in humans (cross-species transmission). Successive spillovers result in a limited chain of transmission in the affected population. If the epidemic ends spontaneously it is referred to as an outbreak.
However, in some rare cases, spillover or successive spillovers lead to a local chain of transmission. The microorganism gradually finds means of adapting and developing in humans. This phenomenon, known as cross-species transmission, or host jump, is where a pathogen becomes established in a new species. The spread of infections from animals to humans is referred to as zoonosis and causes 60% of human diseases according to WHO. The outcome of the infection depends on its transmissibility within the new population.
Stage 3 : circulation

Stade 3 : The epidemic becomes established or even turns into a pandemic through highly efficient transmission within the new host population.
Transmission becomes widespread and an epidemic takes hold due, in particular, to the pathogen's ability to overcome resistance through secondary mutations. Transition of an outbreak to a pandemic is dependent on geographical spread and the basic reproduction number.
The species barrier is normally robust, and although critical, the phenomenon is rare and complex. However, it is possible to pinpoint certain factors that increase the likelihood of occurrence.
The first set of factors relates to biology: microbes adapt in their attempts to thwart tactics used by immune systems. Moreover, hosts' susceptibility to infectious agents varies based on genetic, environmental and social factors. We should also not underestimate the ability of certain animal vectors such as arthropods and mosquitoes to effectively host and then transmit viruses to humans, acting as a link between species, as observed with Zika virus in America in 2016.
The second set of factors relates to human, socio-economic and political issues. The boom in international travel and trade, land use changes and ecosystem deterioration, social inequality particularly with regard to education, and situations involving armed conflict, food insecurity and poor governance are all factors providing favorable conditions for these emerging diseases and determining their transmissibility or failure. Recent examples such as the Nipah virus (in Malaysia in 1999) and SARS-CoV-1 (in China and South-East Asia in 2002-2003) clearly demonstrate how a combination of environmental factors (deforestation, habitat loss for animal reservoirs prompting contact with animal vectors) and social factors (consumption of animal vectors) pave the way for cross-species transmission.
Finally, physical factors linked to the increasing frequency of extreme natural phenomena (floods, earthquakes, storms, fires) are changing the balance of ecosystems and disrupting species interactions.
It would be excessively simplistic to identify human impact on the environment as the sole source of emerging diseases. However, it is clear that human-induced changes to natural habitats increase the risk of contact between humans and animal vectors which are reservoirs of infectious agents. This is further compounded by social factors including the boom in international trade which provides the ideal conditions for global expansion of local phenomena.
Can the next emergence be predicted?
We are well aware of the importance of detecting the potential emergence of a disease at the spillover stage. While predicting such phenomena is as complex as the interlinked factors involved, three simple principles may be applied: monitoring, assessment and reporting. Scientific cooperation is more crucial in this field than in any other, as are modeling, sequencing and diagnostic capacity. The challenges faced are cross-disciplinary and require integrated approaches. Although the SARS-CoV-2 pandemic severely tested our systems, they ultimately proved effective, with the virus sequenced in less than three weeks, diagnostic testing rolled out in less than a month, and a vaccine developed in the space of a year. These technical and technological feats should not overshadow the risk we run and the need to guard against it. In particular, we must consider factors that foster the emergence of new diseases by questioning our interaction with ecosystems, as this is where lies the "genius of infectious diseases" in the words of Charles Nicolle, a French physician and microbiologist trained at the Institut Pasteur and laureate of the 1928 Nobel Prize in Medicine. Although difficult to predict with current scientific resources, it is vital that we urgently detect and report any emergence events in order to nip outbreaks in the bud. Monitoring the environment and animal and human populations forms part of the One Health approach of "one health for humans and animals in one and the same environment."
 Interaction between a dendritic cell and a lymphocyte. |
With Philippe Sansonetti, microbiologist, professor at the Collège de France, holder of the Chair in Microbiology and Infectious Diseases, and professor at the Institut Pasteur.
"There will therefore be new diseases. That is a fatality. (…)"


A woman in pursuit of HIV for over 30 years
Michaela Müller-Trutwin considers her encounter with the virus to which she has dedicated her career as the result of a quest to make herself useful. From University of Bonn lecture halls to "Pasteur" laboratories, first in Bangui then in Paris, she has been researching HIV (the human immunodeficiency virus responsible for AIDS) for over 30 years and has always gone where the need for research has been greatest.
"It all stems from personal commitment," she tells us. While studying biology in Germany, she decided to focus her efforts on countries in the southern hemisphere where needs are most critical, and settled on the Institut Pasteur in Bangui in the Central African Republic. That was in the early 1990s when the AIDS epidemic was raging in Africa. The HIV variants circulating there appeared very different from those circulating in the United States and Europe. Michaela therefore set out to identify the variants circulating in central Africa. However this was no easy task: "The PCR tests available were calibrated for American and European variants and sometimes proved ineffective", she explains. It took her a year to implement the first PCR tests for HIV-related viruses in monkeys living in the Central African Republic. Her research, enhanced by a bioinformatics-based approach, proved crucial in more effectively tracking down circulating variants and helped advance vaccine studies. She also investigated the origin of the virus and, in collaboration with the Pasteur Center in Cameroon and a Paris Public Hospital Network (AP-HP) team, she characterized the first SIV (the primate equivalent to HIV) related to HIV-1 in chimpanzees in south-eastern Cameroon.
After joining the Institut Pasteur laboratory in Paris led by Françoise Barré-Sinoussi, laureate of the 2008 Nobel Prize in Medicine with Luc Montagnier for their joint discovery of HIV, she continued to work closely with the Pasteur Network (Bangui, Dakar and Cameroon) and the International Center for Medical Research in Gabon. Skills and knowledge transfer was a key part of her mission.
While researching African green monkeys infected with SIV from very close quarters in the early 2000s, she noted their failure to develop the disease and attempted to pinpoint the mechanism that conferred natural protection against AIDS. "We discovered that they are able to control inflammation resulting from the infection, and helped to determine that inflammation plays a major role in immunodeficiency and the development of AIDS in humans." This discovery of rapid inflammation resolution helped highlight the importance of minimizing time from infection to triple therapy initiation.
However, Michaela did not stop there. She continued her research on the green monkey model and in 2021 discovered the hitherto unknown role of natural killer cells (see p. 36) in controlling viral replication and reducing hidden viral reservoirs in our lymph nodes. This remarkable breakthrough opens up new avenues to find a cure for HIV. One thing is certain: Michaela may not yet be done with HIV but her quest to make herself useful has already been accomplished.
The COVID-19 pandemic in three acts
Prologue
Much has been said and written about the COVID-19 pandemic. Although we still lack the necessary scientific hindsight, it is not the first pandemic the modern world has had to face. Many experts predicted that the risk of an emerging disease with pandemic potential would come from a respiratory virus inherently more transmissible. This one nevertheless outpaced existing alert and response systems, proving the need for a highly connected global scientific community. Here is a look back at the pandemic in three acts.
Act I: identifying the threat
 Sample of bronchial cells grown in culture and stained blue. In orange, the SARS-CoV-2 coronavirus. |
Fact. Research published in the journal Nature by Pasteur Network teams in Paris and Laos in early 2022 identified a coronavirus in bats in Laos with a human ACE2 receptor binding domain (RBD) that was virtually identical to the SARS-CoV-2 RBD. Bats appear to be the reservoir for the virus but the intermediate host between bats and humans is still unknown, although pangolins were suspected at one point. While determining its origins was important for preventing reintroduction of the virus into the human population, identifying the infectious agent was a matter of public health urgency.
With high throughput sequencing, it is now possible to determine the genome of an unknown pathogen in 2 to 4 weeks. The first whole-genome sequence of SARS-CoV-2 was published on January 12, 2020, which was a mere 12 days after the first cases were reported and a testament to the Chinese scientists' strong response.
Once the sequence of a pathogen has been characterized, a diagnostic test can be developed to identify the first outbreak clusters and imported cases, and to triage suspected patients. The Institut Pasteur in Paris and its CNR for Respiratory Infection Viruses undertook this task and produced the first testing kits based on the RT-qPCR technique on January 31, 2020. The kits were immediately distributed to hospitals.
The first cases had reached France just days previously on January 24, 2020 and the virus was isolated at the Institut Pasteur on January 27. The outbreak then gained pace and all the machinery was set in motion to monitor its development.
Act II: understanding and responding to the outbreak in real time
WHO declared the outbreak a pandemic on March 11, 2020. Escalation to pandemic status was determined by the numbers of cases and their geographical spread. By now the virus was circulating on all continents and nothing seemed to halt its progress.
A task force was appointed at the Institut Pasteur on January 22 with weekly meetings to coordinate overall and multidisciplinary research efforts. On February 12 and March 7, 2020, two calls for research proposals were issued within the Pasteur Network, which includes all members of the international network.
All expertise was focused on the pandemic. Each department forged its own approach to tackling the issue within its given remit to deepen understanding of the outbreak unfolding before our very eyes. This massive response and collaboration within the global scientific community was on a virtually unprecedented scale.
In Paris, work began on epidemiological studies of places of SARS-CoV-2 transmission, mathematical modeling of outbreak dynamics, and development of diagnostic and therapeutic approaches – in 2021, this included monoclonal antibody and vaccine therapy with progress made on a potential intranasal vaccine developed with TheraVectys resulting in two publications demonstrating its efficacy in animal models which should enter clinical trials by late 2022, and previously in 2020 two other avenues of investigation: a measles vector-based vaccine candidate, which proved insufficiently immunogenic and was discontinued, and a DNA vaccine (see 2020 annual report). Scientific knowledge was useful for monitoring the outbreak and advising the authorities on public health response measures. Nearly a hundred research projects were initiated at the Institut Pasteur in the first months of the outbreak to increase understanding and drive response.
Act III: a return to long-term scientific research
Although pandemics always come to an end, it is difficult to predict their outcome in advance. How and when will we emerge from this one? With each new emergence the same questions arise. This is perhaps because several timelines coexist. First, there is the short-term public health timeline which occurs in near real time and gives rise to the resources needed for patient management and outbreak response. Then there is the long-term timeline of scientific research aimed at elucidating the biology and emergence of SARS-CoV-2 through a multidisciplinary approach. In 2021, the Institut Pasteur launched collaborative projects, each involving at least 4 of its research teams, to address key scientific questions regarding this pandemic. The projects will span several years and their findings should provide insights into the SARS-CoV-2 virus and its mutations, which are a source of concern for the coming months, and other coronaviruses, and so expand our knowledge to limit new emergences in the future.
Initial lessons and prospects
The COVID-19 pandemic is not over yet. Research and efforts to discover new ways to control the disease must continue. For instance, clinical trials are planned for the monoclonal antibody developed with SpikImm – see p.47. The pandemic has also reminded us that outbreaks of this scale can still occur and cannot yet be consigned to the history books. It confirms the epidemic potential of respiratory viruses and the need for vigilance. Moreover, it calls for a concerted effort, both in- and outside the scientific community, to identify and control factors precipitating the emergence of diseases, in particular climate change and human-animal interactions.
The pandemic has also highlighted the need for the Institut Pasteur to build its capacity in response to emerging diseases, especially within the field of vaccinology. This will be a key challenge for future years. On scientific issues, researchers have remained highly connected, sharing scientific knowledge in real time, as well as scientific publications through preprints with commentary on social media platforms such as Twitter. However, this raw scientific data provided via platforms that was inherently complex and in need of peer review required constant monitoring to maintain its quality for the benefit of global health.
With Arnaud Fontanet, head of the Institut Pasteur Epidemiology of Emerging Diseases Unit and co-head of the emerging infectious diseases priority area (2019-2023 Strategic Plan).
- Elucidating the biology of the virus through genetic engineering
- Understanding long COVID
- Examining virus interaction with target cells using structural imaging technology
- Examining SARS-CoV-2 infection through animal imaging
- Examining virus interaction with host cells
- Developing therapeutic strategies
- Examining immune response to infection and vaccination
Shigellosis, profile of an emerging disease
Shigellosis is a diarrheal disease endemic in several regions of the world. Outbreaks occur during mass gatherings in environments with inadequate hygiene conditions.
The disease is caused by an intestinal bacterial pathogen for which humans are the sole reservoir. It has considerable infectious potential, with only 10 to 100 bacilli required to cause the disease. With the emergence of antibiotic-resistant strains and morbidity due to the long-term consequences of recurrent infections it is under special surveillance.
Origins and emergence
- Shigella bacteria, specialized clones of the Escherichia coli species, discovered in 1897 by the bacteriologist Kiyoshi Shiga.
- 4 species: Shigella dysenteriae, Shigella flexneri, Shigella boydii and Shigella sonnei. S. sonnei is the most prevalent species in France (70% of cases) and is an emerging pathogen reported worldwide. S. flexneri causes two-thirds of shigellosis cases in Africa and Asia.
Transmission
Fecal-oral transmission via water or food contaminated by infected feces or flies, and also by infected individuals to their close contacts.
Surveillance
The Institut Pasteur is responsible for monitoring cases of shigellosis in France (both mainland and overseas) through the Enteric Bacterial Pathogens Unit accredited as a National Reference Center (CNR) for Escherichia coli, Shigella and Salmonella.
Developments
- Vaccine: successful Phase I clinical trial in 2021 for a conjugate vaccine candidate derived from synthetic sugars developed by the Institut Pasteur (Chemistry of Biomolecules/Molecular Microbial Pathogenesis units) for the prevalent S. flexneri serotype (see p. 29). Phase IIa clinical trial underway in Kenya to establish the efficacy and safety of the monovalent vaccine candidate in the target population of very young children. The results are expected in 2023.
- Identification: update of methods for identifying and typing Shigella strains using a bacterial genome sequencing approach (Institut Pasteur Enteric Bacterial Pathogens Unit).
- Diagnostics: dipstick tests developed for prompt 15-minute diagnosis (Pasteur Network and Military Health Department).
Humans, the environment, animals and diseases

The COVID-19 outbreak has forced us to face a simple truth: life is unique. The concept of global health has gained ground throughout the world, both within WHO (World Health Organization) and the Institut Pasteur. If the environment is suffering, how can animals thrive? And within the animal kingdom, how can we humans be the only ones to stay healthy? How can we protect the oceans if we continue to dump our waste in the rivers? The idea of impermeable boundaries between partners of the living world is a misconception, and one that is potentially fatal. Any attack on life is made because other living beings have something to gain. This is probably the first of Louis Pasteur's legacies.
Before Pasteur, medicine was primarily based on observation. Thanks to him and others, we have made progress on discovering causes. It is astonishing to note that lethal infectious diseases are caused by tiny living beings, or even tinier inert particles consisting of a single genome (DNA or RNA) and an envelope – viruses. This crisis reminds us of our vulnerability. The more connected the world is, the more dependent we are on those things that seem most insignificant within it. We are far more dependent on the weakest elements than the strongest ones. Threats are more likely to emerge from the weakest elements. My work on the "geopolitics of mosquitoes" and swine fever has given me a little more insight into the workings of epidemics. Will we learn the lessons of this latest health crisis?
This crisis also forces us to consider our lifestyles. The way we produce food has been a major factor in recent emergences. I have a long-standing interest in agriculture and particularly the transitions required in this area. Food prices are subject to incredible downward pressure, with the proportion of household spending on food falling from 30% to 10% in a very short space of time. Some countries such as China are restructuring their capacities with multi-story farms including units of 28,000 sows spanning nine floors in one building and producing on an industrial scale. How can we think for one second that these systems will guarantee our health and safety?
The health crisis and climate crisis amount to the same challenge. Like life itself, crises are connected. Every year, parasites carried by mosquitoes claim over 700,000 lives. And we couldn't care less because it mainly affects poor countries. Yet I certainly do not view the past through rose-tinted spectacles. Globalization has pulled hundreds of thousands of human beings out of poverty, but it has also come with some horrific epidemics. Our opportunity lies in admitting the need for change.
Unity of life, global economy, planetary connection... How do we reconcile this interconnectedness with an increasingly unequal society? The more devastating a crisis, the harder the poorest are hit. We are faced with a huge challenge. However, with the right energy, we can regain control of our future. Solutions are out there that all require research. In such a context, who would be foolish enough to cut research budgets?
Extract from Erik Orsenna's "Tract de Crise" [Crisis Tract] published by Gallimard in March 2020. Text written by the author based on an interview with Fabrice Moyon for the March 21, 2020 issue of Ouest-France newspaper and adapted for this annual report in April 2022.






