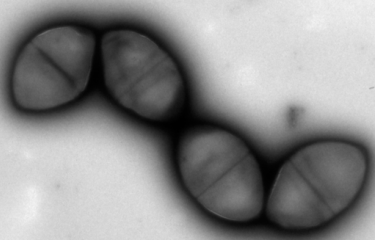
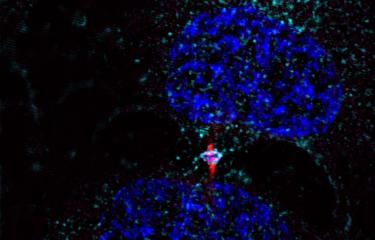
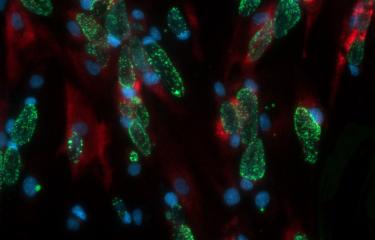
Scientists at the Institut Pasteur have developed a method to isolate midbodies, bridges that link cells undergoing cytokinesis. Using the latest mass spectrometry methods, they produced a comprehensive inventory and revealed the presence of 1,732 proteins! By characterizing three of these proteins in detail, they pinpointed novel molecular mechanisms involved in both cell division and virus budding.
The Membrane Traffic and Cell Division Unit in the Institut Pasteur's Department of Cell Biology and Infection is investigating a question that is fundamental for our understanding of biology: how does a cell produce two identical daughter cells? In particular, how does a parent cell physically cut itself in two to form two independent daughter cells (a process known as cytokinesis)?
Back in 1891, leading German zoologist Walther Flemming described how cells undergoing cytokinesis were linked by an intercellular bridge with a morphologically unique structure at its center known as the midbody or Flemming body. For the past decade, scientists have been aware that the midbody is a key platform for the recruitment of all the proteins responsible for the process of abscission, when the parent cell is finally cleaved in two. Identifying the proteins in the midbody is therefore a vital stage in revealing novel molecular mechanisms responsible for abscission, which remain largely unknown.
A method for isolating midbodies
The Membrane Traffic and Cell Division Unit recently developed a method to isolate and obtain highly pure, intact midbodies from human cells in culture and to produce an exhaustive inventory of them using the latest mass spectrometry methods. This novel inventory, named the Flemmingsome in honor of Walther Flemming, reveals the presence of a surprisingly high number of proteins in the midbody (1,732). Of these proteins, 489 are found enriched in the midbody, more than 300 for the first time, all representing new candidates for abscission. Three proteins (Syndecan-4, Syntenin and ALIX) were characterized in detail, revealing the first membrane protein directly involved in cytokinesis.
This research was conducted in conjunction with other research teams at the Institut Pasteur: the Flow Cytometry Platform (development of the purification protocol), the Proteomics Platform (development of new methods for isolating peptides), the Bioinformatics and Biostatistics Hub (statistical analysis and creation of a dedicated website) and the Imaging Platform (electron microscopy and super-resolution microscopy).
Advances in basic cell biology and new avenues for virology
The research also unexpectedly revealed similar molecular mechanisms implicated in both the final stages of cell division and the shedding of some enveloped viruses (like HIV, Ebola or HBV) from infected cells. These advances therefore open up new avenues for both basic cell biology and virology, by proposing novel targets whose inhibition could potentially disrupt viral propagation.




Source
The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis, Nature Communications, April 22, 2020.
Cyril Addi1,2, Adrien Presle1,2, Stéphane Frémont1, Frédérique Cuvelier1, Murielle Rocancourt1, Florine Milin1, Sandrine Schmutz3, Julia Chamot-Rooke4, Thibaut Douché5, Magalie Duchateau5, Quentin Giai Gianetto5,6, Audrey Salles7, Hervé Ménager6, Mariette Matondo5, Pascale Zimmermann8,9, Neetu Gupta-Rossi 1,10 & Arnaud Echard 1,10
1. Membrane Traffic and Cell Division Lab, Institut Pasteur, UMR3691, CNRS, F-75015 Paris, France.
2. Sorbonne Université, Collège doctoral, F-75005 Paris, France.
3. Institut Pasteur, UTechS CB, F-75015 Paris, France.
4. Institut Pasteur, Mass Spectrometry for Biology Unit, C2RT, USR 2000, CNRS, F-75015 Paris, France.
5. Institut Pasteur, Proteomics Platform, Mass Spectrometry for Biology, C2RT, USR 2000, CNRS, F-75015 Paris, France.
6. Hub de Bioinformatique et Biostatistique – Département Biologie Computationnelle, Institut Pasteur, USR 3756 CNRS, F-75015 Paris, France.
7. UTechS Photonic BioImaging PBI (Imagopole), Centre de Recherche et de Ressources Technologiques C2RT, Institut Pasteur, Paris 75015, France.
8. Centre de Recherche en Cancérologie de Marseille (CRCM), Equipe labellisée Ligue 2018, Aix-Marseille Université, Inserm, CNRS, Institut Paoli Calmettes, 13009 Marseille, France.
9. KU Leuven, Department of Human Genetics, University of Leuven, B-3000 Leuven, Belgium.
10. These authors jointly supervised: Neetu Gupta-Rossi, Arnaud Echard.