In an article published in PLoS Genetics on July 5, scientists from the Institut Pasteur and the Belgian-based Université catholique de Louvain identify for the first time the genetic and metabolic mechanisms underpinning the therapeutic action of a bacteriophage known for its therapeutic potential. Given the worrying rise in bacterial resistance to antibiotics and the difficulties in developing effective new molecules, there has been renewed interest within the scientific community in recent years in phage therapy, which makes use of these bacterial viruses. This discovery will contribute to ongoing efforts to develop this treatment, which has been largely neglected in Western countries.

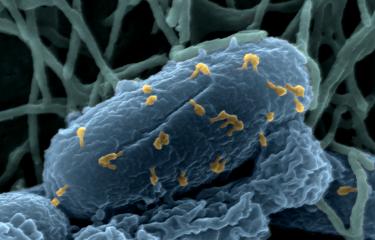
Bacteriophages on the Escherichia coli bacterium © Institut Pasteur
Phage therapy is a medical approach for the treatment of bacterial infectious diseases which makes use of the natural ability of some viruses, known as bacteriophages, to kill specific bacteria that they recognize. Phage therapy was first proposed nearly a century ago at the Institut Pasteur by the French-Canadian scientist Félix d'Herelle. Although the technique was initially used worldwide, from the 1940s onwards it was particularly developed in Eastern Europe, while Western countries gradually lost interest in the approach and preferred to focus their attention on antibiotics, thought to be more promising. Medical use of phage therapy is currently hindered by the viral nature of this antibacterial treatment and the lack of precise knowledge about how bacteriophages work.
However, given the rise in bacterial resistance to antibiotics and the worrying prospect of a potential therapeutic impasse, scientists are once again beginning to explore the possibilities of bacteriophages. This field has been the focus of research by scientists at the Institut Pasteur, led by Laurent Debarbieux[1], and the Université catholique de Louvain in Belgium, led by Rob Lavigne. Their long-awaited findings, published today, shed light at molecular level on the original action mechanisms used by a specific bacteriophage, known for its therapeutic potential, as it infects the opportunistic pathogenic bacterium Pseudomonas aeruginosa.
The scientists' research reveals that the bacteriophage's entire infection strategy, from hijacking the cell to implementing its own proliferation program, is based on RNA metabolism, a fundamental mechanism in the biological production process.
The scientists demonstrated that during its infectious cycle, the bacteriophage is capable of drastically destabilizing the cell by impairing its RNA at an early stage. By contrast, RNA synthesis in the bacteriophage is particularly active, reflecting the high level of activity associated with its own proliferation.
The scientists also showed that the bacteriophage makes use of control mechanisms involving RNA fragments known as "small RNA" and "antisense RNA" to meticulously orchestrate the various stages in the production of new viral particles.
This research emphasizes the potential value of characterizing bacteriophages with therapeutic properties, both in terms of the contribution to fundamental research and also to improve understanding of the action mechanisms behind the antibacterial activity of bacteriophages. "This research should encourage further characterization of other bacteriophages with therapeutic potential," comments Laurent Debarbieux. "We hope that it will ultimately pave the way for the use of these viruses for medical purposes in France."
Source
Next-Generation“-omics”Approaches Reveal a Massive Alteration of Host RNA Metabolism during Bacteriophage Infection of Pseudomonas aeruginosa, Plos Genetics, July 5, 2016.
Anne Chevallereau (1,2☯),Bob G. Blasdel (3,☯),Jeroen De Smet (3), Marc Monot (4), Michael Zimmermann (5), Maria Kogadeeva (5), Uwe Sauer (5), , Peter Jorth (6), Marvin Whiteley (6), Laurent Debarbieux (1)*, Rob Lavigne (3)*
1) Institut Pasteur, Molecular Biology of the Gene in Extremophiles Unit, Department of Microbiology, Paris, France,
2) Université Paris Diderot, Sorbonne Paris Cité, Cellule Pasteur, Paris, France,
3) Laboratory of Gene Technology, Department of Biosystems, KU Leuven, Leuven, Belgium,
4) Institut Pasteur, Laboratoire Pathogenèse des bactéries anaérobies, Département de Microbiologie, Paris, France,
5) Institute of Molecular Systems Biology, Eidgenössische Technische Hochschule (ETH) Zürich, Zürich, Switzerland,
6) Department of Molecular Biosciences, Institute for Cellular and Molecular Biology, Center for Infectious
Disease, University of Texas, Austin, Texas, United States of America
☯Contributions égales de ces auteurs.