Antibiotics: when bacteria fight back
Who hasn't taken antibiotics at least once in their lifetime? It is clear that these drugs, which combat a wide range of bacterial infections including pneumonia, bronchitis, ear infections, meningitis, urinary tract infections and septicemia, save millions of lives. But their efficacy is under real threat, and the World Health Organization warns that "one day no antibiotics may be left to treat common bacterial infections".
Those infections would once again become life threatening, and society would return to the pre-antibiotic era. It is currently possible to die from an apparently "commonplace" infection if it is caused by antibiotic multidrug-resistant bacteria. In Europe, 400,000 people are infected by these bacteria every year, and 25,000 die from their infections. Increasing antibiotic resistance will lead to a dramatic rise in these figures, as predicted in the report by Lord J. O'Neill on the impact of antimicrobial resistance by 2050.
Antibiotics used from the 1940s
To shed some light on the alarming problem of antibiotic resistance, let's start at the beginning. The first antibiotic, penicillin G, was discovered in 1928 by the Scottish biologist Alexander Fleming, but it was only used from 1941 onwards (see below). In the meantime, another class of antibiotics, sulfonamides, whose action was revealed by Institut Pasteur scientists, began to be widely used and went on to save thousands of lives during the Second World War.
Today, 70 years after antibiotics first became available, there are more than 15 antibiotic families, which differ in terms of their chemical structure and their mechanism of action against bacteria. The problem of antibiotic resistance was raised by Fleming himself back in 1945. He foresaw the risks associated with improper use of the molecule that he had discovered, predicting that it would lead to a situation in which, instead of eliminating the infection, "the microbes are educated to resist penicillin and a host of penicillin-fast organisms is bred out which can be passed to other individuals and perhaps from there to others until they reach someone who gets a septicemia or a pneumonia which penicillin cannot save."
The accidental discovery of penicillin...

September 3, 1928: Sir Alexander Fleming, a biologist well known for his scientific talents but also for his messy laboratory (according to what is commonly said) at St Mary's Hospital in London, came back from holiday to find that a forgotten culture of staphylococci, bacteria whose properties he was studying, had been contaminated by mold. When he observed it, he noticed that not a single colony of bacteria remained in the area where the mold had developed, whereas the bacteria further away in the Petri dish were still there. Could this fungus have the power to kill bacteria? Alexander Fleming identified the mold as Penicillium notatum. He isolated an extract and gave the agent the name "penicillin". He then investigated its effects and noticed that it acted not only on staphylococci but also on many other bacteria, including those responsible for scarlet fever, diphtheria, pneumonia and meningitis.
The monumental importance of his discovery was only understood later, when other researchers - Howard Walter Florey and Ernst Boris Chain - carried out work to purify penicillin G and produce it in sufficient quantities to treat human infection, leading to its use in medical applications from 1941 onwards. They shared the Nobel Prize in Medicine with Alexander Fleming in 1945.
The end of the golden age of antibiotics
Fleming's warning was not initially heeded, and antibiotics were used extensively from the 1940s onwards: they were "magic bullets" which, together with a number of vaccines, brought about a huge reduction in the impact of bacterial infectious diseases, at least in industrialized countries. But the "golden age" of antibiotics came to an end in the early 1990s, when it became ever clearer that a worryingly high – and still growing – number of bacteria had become resistant to antibiotics, and that the discovery of new molecules to fight against resistant bacteria was drying up.
Doctors were increasingly finding themselves in a therapeutic impasse when treating some patients since there were no antibiotics that had even the slightest impact on their infection. And the phenomenon is gaining pace. The situation has become particularly critical in hospitals, where it is only natural that large numbers of antibiotics are used to treat patients: half the human consumption of antibiotics in France takes place in hospitals. This "drug pressure" causes resistance, and resistant bacteria spread easily from one patient to another, infecting them all the more readily as they are already in a weakened state. In hospitals, more than one-fifth of Staphylococcus aureus bacteria are resistant to methicillin and other antibiotics of the family of penicillins used to treat S. aureus infections (such as pulmonary and bone infections, septicemia, etc.). A large number of germs responsible for hospital-acquired infections (also known as nosocomial infections) are becoming increasingly resistant to several antibiotics.
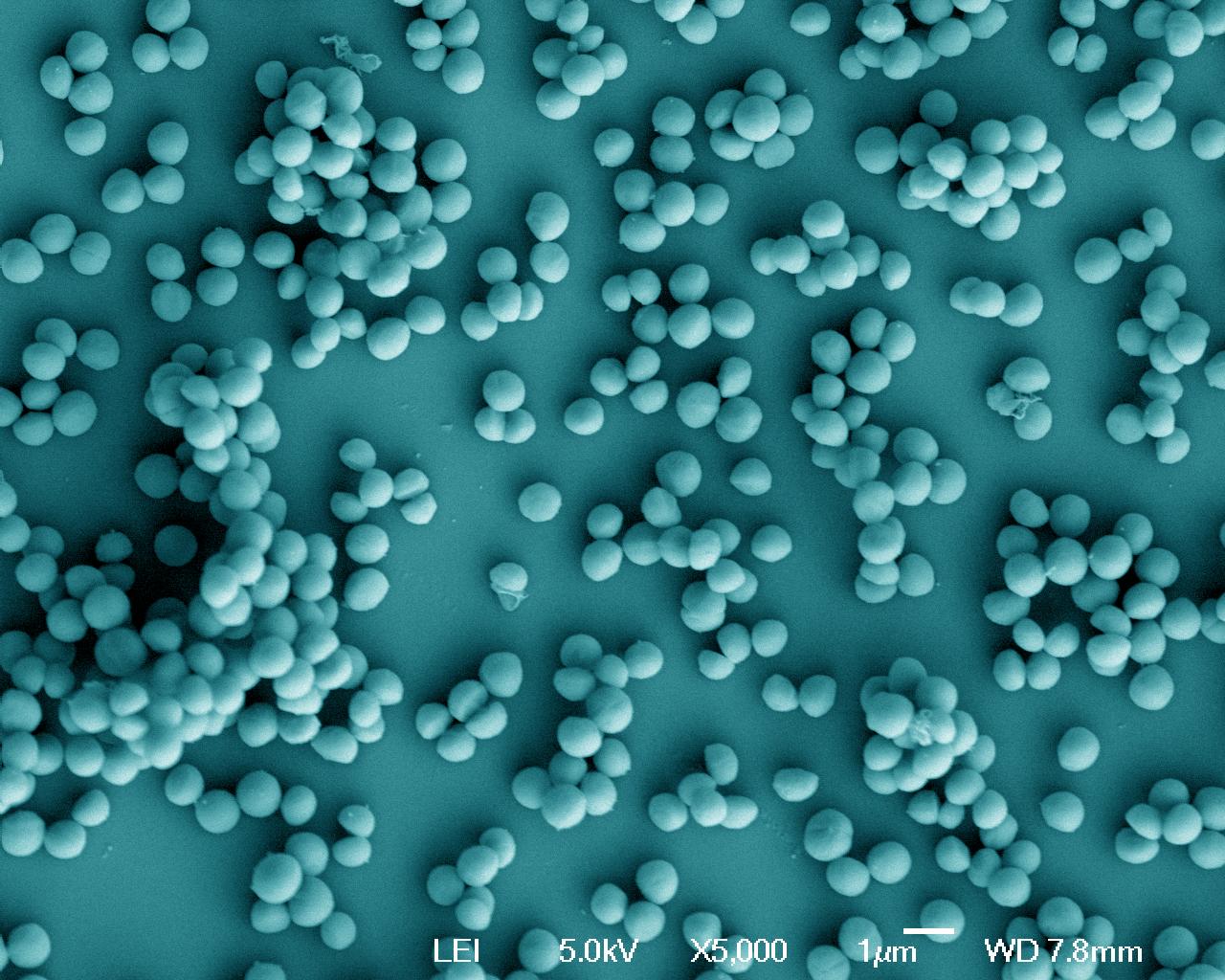
Staphylococcus aureus, grown in a rich environment. Scanning microscopy. Copyright: Institut Pasteur/Adeline Mallet, plateforme de microscopie ultrastructurale - Unité Biologie des bactéries pathogènes à Gram-positif
An everyday concern
The problem of antibiotic resistance is no longer limited to the hospital environment; it has now spread to community-acquired infections. On medical website forums, those affected are beginning to raise questions, such as this woman who describes her own experience: "For the past six months I have been suffering from urinary tract infections, always caused by an Escherichia coli germ resistant to several antibiotics." She describes the new treatment recommended by her family physician, based on a new antibiotic, and wonders: "Won't this highly resistant germ develop even greater resistance? What do you think?" Antibiotic resistance has now become a daily reality affecting many patients – and it goes without saying that it is a major concern for both doctors and scientists (See the interview with Philippe Glaser below).

One of the consequences of antibiomicrobial resistance is the delay in giving effective treatment to the patient. An important avenue of research is the development of a rapid test to ensure that medical staff can prescribe an antibiotic that will be active against a given infection.
What is your view on the phenomenon of antibiotic resistance today?
Antibiotic resistance is a public health issue that is now being addressed by public authorities, especially in hospitals. But treating patients carrying multidrug-resistant bacteria in hospitals is a costly endeavor – not only in financial terms, requiring the allocation of dedicated staff to prevent the transmission of these bacteria to other patients and avoid outbreaks, but also in psychological terms for the patients concerned, as they need to be given stringent monitoring. But thanks to these efforts, the level of resistance is under control in France and in most European countries – unlike in some other nations around the world, especially low-income countries. Antibiotic resistance can be considered as an "emerging disease", since resistant bacteria are spreading across the globe. To combat the phenomenon of antibiotic resistance, we need to adopt a comprehensive, global view.
What causes antibiotic resistance?
Bacteria are capable of acquiring genes from resistant bacteria that confer resistance to antibiotics. A given individual may be a carrier of a resistant bacterial strain in the gut microbiota without experiencing any health problems or apparent symptoms. All it takes is for this individual to be infected by pathogenic bacteria, and the resistant strain can transfer its resistance genes to these new bacteria. The phenomenon may occur quickly and may even be aided by antibiotics.
What can we do to combat antibiomicrobial resistance?
One of the consequences of antibiomicrobial resistance is the delay in giving effective treatment to the patient. Indeed, identifying antibiotic resistance in bacteria responsible for an infection is currently a lengthy process which only occurs in the event of therapy failure. An important avenue of research is the development of a rapid test to ensure that medical staff can prescribe an antibiotic that will be active against a given infection.
Resistant bacteria are emerging in various places worldwide, and measures need to be taken at global level, perhaps coordinated by the World Health Organization (WHO). At our level, we can try to minimize the spread of these bacteria by enforcing the surveillance and hygiene measures that have already been adopted in France, in both hospitals and community settings. We also need to make more judicious use of antibiotics: the right dose, the right treatment period, and the right combination. And this applies not only to humans but also to animals, so that we can curb resistance reservoirs.
What research is being carried out at the Institut Pasteur?
Current research programs at the Institut Pasteur combine global epidemiology (via the laboratories in the Institut Pasteur International Network), genomics and bioinformatics to describe these phenomena and unravel the underlying mechanisms. Scientists are also investigating how bacteria exchange resistance genes so that they can develop strategies to block these transfers. To find new antibiotics, Institut Pasteur teams are investigating the biosynthesis of the bacterial envelope, a target of many antibiotics. This knowledge will serve as a basis for the identification of new active molecules, either in molecule libraries containing a wide range of molecular structures, or among natural products. The Institut Pasteur is also developing a number of alternative strategies to tackle multidrug resistant bacteria, including phage therapy, antimicrobial peptides and the possibility of "hijacking" the CRISPR system, a bacterial immune system, to specifically kill resistant bacteria.
Photo: Philippe Glaser, head of Ecology and Evolution of Antibiotics Resistance Unit. Copyright: Institut Pasteur - photo François Gardy
"Antibiotics are not automatic!"

Public health authorities are taking action. People living in France will no doubt remember the "Antibiotics are not automatic!" campaign launched in 2002 by the French health insurance authority. The idea was to raise awareness of the fact that antibiotics are only useful in treating bacterial infections; they have no effect on viral infections, for example those often responsible for seasonal conditions such as colds, sore throats and bronchitis. The campaign had a considerable impact on antibiotic consumption in France, which fell by 15%. In 2010, public authorities launched a new campaign which tackled the question of antibiotic resistance in France even more directly, with the slogan "if we misuse them, they will become less effective". Since then, the French health insurance authority has continued its efforts to raise awareness about using antibiotics correctly (page in French).
The promise of nanodrugs and the search for new antibiotics
"Using nanoparticles – assemblies of sugars, fats and polymers – to deliver antibiotics could enable us to administer them in smaller doses, or to employ antibiotics that are currently no longer in use because they are poorly tolerated," explains Brigitte Gicquel, Head of the Mycobacterial Genetics Unit at the Institut Pasteur. The tuberculosis specialist is coordinating the European project NAREB,* launched in February 2014, in which 14 laboratories are working to develop nanodrugs to treat two types of infection that are a major cause for concern: methicillin-resistant Staphylococcus aureus (MRSA), frequently responsible for severe hospital-acquired infections, and multidrug-resistant tuberculosis. Research by the NAREB consortium has shown for a small number of antibiotics that it is possible to associate them with nanoparticles without affecting their activity in vitro and in vivo. The scientists are now working to demonstrate the superiority of an antibiotic-nanoparticle combination that could change currently used formulations.
Multidrug-resistant tuberculosis currently represents 3.7% of new tuberculosis (TB) cases worldwide and 20% of previously treated cases, further complicating treatment of a condition that requires a lengthy drug regimen (several months with at least 4 antibiotics for "drug-susceptible" TB).
What's more, some 84 countries have already reported cases of "extensively drug-resistant tuberculosis" (XDR-TB). "Given this dramatic situation and the disengagement of most industry stakeholders, we as academic laboratories must make an effort to find new antibiotics and new formulations," emphasizes Brigitte Gicquel, whose team is actively involved in the research efforts to develop new drugs that are currently taking place within multi-institution consortia such as NAREB. Whether nanodrugs or new antibiotics, new solutions are urgently needed given the growing resistance of the tuberculosis bacillus. TB, which was responsible for one in seven deaths in Europe in the 19th century, remains a global health concern, with 8.5 million new cases and 1.3 million fatalities every year worldwide.
* Nanotherapeutics for antibiotic resistant emergent bacterial pathogens.
Investigating the impact of small doses

What impact do weak doses of antibiotics have on the emergence of antibiotic resistance? "Even at antibiotic concentrations a hundred times lower than those capable of killing bacteria, a bacterial stress response known as the SOS response is triggered, promoting the acquisition of resistance genes," replies Didier Mazel, Head of the Bacterial Genome Plasticity Unit at the Institut Pasteur. In 2013, the scientist demonstrated this phenomenon in several pathogenic bacteria including Vibrio cholerae, the cholera agent, and Klebsiella pneumoniae, responsible for respiratory infections. For him, "this observation particularly points to the problem of low concentrations of antibiotics found in wastewater or rivers: the environment could play a non-negligible role in the development of antibiotic resistance."
These findings paved the way for other fundamental research. Recently, Didier Mazel and Zeynep Baharoglu, a scientist in his unit, identified the entry pathway of a family of antibiotics into Enterobacteriaceae, a discovery that could lead to the development of methods to improve bacterial absorption of these antibiotics. So there are real hopes for the future of antibiotic treatment in human medicine, whether by improving how we use them, boosting their efficacy or reducing their side effects.
Limiting the damage
The aim now is to restrict antibiotic consumption so as to curb the spread of resistance. The emergence of antibiotic resistance is a natural biological phenomenon that occurs when bacteria employ mechanisms to resist being "attacked" by antibiotics, such as mutation or the acquisition of resistance genes from other already resistant bacteria. Misuse of antibiotics in humans and animals (see below) – more than half the antibiotics produced in the world are consumed by animals, according to WHO – is accelerating the process. Another major difficulty is limiting the spread of resistant bacteria, which travel via humans. Some of these bacteria have hit the headlines, such as the various bacterial species carrying the NDM-1 gene (from the enzyme New Delhi metallo-beta-lactamase), which makes them resistant to the very latest antibiotics, carbapenems, used to treat multidrug-resistant infections.
These bacteria were detected in 2009 in a Swedish patient who was admitted to hospital in India, and the following year in the United Kingdom in patients who had made use of "medical tourism" to travel to India or Pakistan for cosmetic surgery.
The consequences of antibiotic use in farming

Antibiotic resistance of bacteria among animals, encouraged by high levels of drug pressure on farms, can contribute to antibiotic resistance observed in humans. Scientists from the Institut Pasteur examined early cases of Salmonella resistance to ampicillin, 45 years after their appearance. In late 2017, they determined precisely how resistance has emerged to this broad-spectrum antibiotic, that is still widely used today.
Ampicillin was the first synthetic penicillin found to be effective in treating Enterobacteriaceae* infections. This broad-spectrum antibiotic, one of the most widespread antibiotics in the world, was released on European markets in 1961. Shortly afterwards (1962-1964), the first outbreaks caused by ampicillin-resistant strains were observed in humans. These outbreaks, caused by the zoonotic bacterium Salmonella enterica serotype typhimurium, particularly affected the United Kingdom.
The very short lapse of time between the release of the antibiotic on the market and the first cases of resistance described in the scientific literature prompted the Institut Pasteur's scientists to investigate the emergence of ampicillin resistance. They found that bacteria that can pass on genes resistant to ampicillin actually emerged, unexpectedly, in the late 1950s. By analyzing the genomes of hundreds of historical samples of Salmonella, they proved that resistance first emerged well before ampicillin was released on the market for human use.
This discovery suggests that low doses of penicillin G, routinely fed to livestock in North America and Europe in the 1950s to boost growth, may have encouraged Salmonella resistant to this new antibiotic to evolve and spread to humans a few years later. The findings were published just weeks after WHO called for an end to routine antibiotic use to promote growth and prevent disease in healthy farm animals.
* Enterobacteriaceae are a large family of Gram-negative bacteria that includes Salmonella, Escherichia coli, Yersinia pestis, Klebsiella and Shigella.

Dr. Francois-Xavier Weill, Head of the Institut Pasteur's Enteric Bacterial Pathogens Unit, who led the study.
Our findings suggest that antibiotic residues in farming environments such as manure, soil and waste water in the 1950s may have had a much greater impact on the spread of ampicillin resistance than previously thought.
Read the press release (in French)
Shared responsibility
How can we deal with these major risks? "WHO calls on all key stakeholders, including policy-makers and planners, the public and patients, practitioners and prescribers, pharmacists and dispensers, and the pharmaceutical industry, to act and take responsibility for combating antimicrobial resistance." Although it is clear that research needs to be stepped up to develop new antibiotics, the problem of bacterial resistance is one that concerns us all, and we should all endeavor to follow some basic guidelines (see below: Antibiotics, a user guide).
Antibiotics, a user guide:
- Never obtain antibiotics without a medical prescription.
- Do not automatically expect your doctor to prescribe you antibiotics every time you have an infection. Viruses are responsible for many respiratory infections, especially colds and flu. Antibiotics only cure bacterial infections; if they are ineffective in treating your condition, there is no point in taking them as they will only make you more vulnerable in future.
- If you are prescribed antibiotics, make sure you follow the exact instructions given by your doctor or pharmacist. Keep taking the antibiotics until the end of the prescribed course, even if you start to feel better. If you fail to do so the infection with resistant bacteria may come back.
- Do not take medication prescribed for another person or another condition.
- Do your best to prevent infections. Wash your hands regularly and encourage your family and colleagues to do likewise.
- Preventing some bacterial and viral infections through vaccination is also an important step.
Source: World Health Organization (WHO)
"Combating Resistance: microbes and vectors"
Considering the threat that microbes and vectors resistance represent at the global scale, the Institut Pasteur has decided to focus the 4th edition of the Institut Pasteur International Network Symposium on these major issues. It will in particular bring together scientists, clinicians and public health experts to:
- Showcase high-level research to identify, monitor or predict the emergence of resistant pathogens and vectors;
- Highlight the current efforts in translational science to develop new tools and technologies necessary to tackle the challenges of diagnostic and surveillance of these health threats;
- Examine innovative solutions to practically develop these tools on the field in resource-limited settings.



