The 3D organization of chromosomes plays a role in many biological processes. The ring formed by the cohesin complex is an important regulator of this organization, especially in mammals where it establishes chromatin loops during interphase. Researchers from the Spatial Regulation of Genomes Unit and the CNRS show that such loops structure yeast chromosomes in metaphase and characterize the proteins regulating their size and position.
The molecular motor cohesin is a ring-shaped complex of the SMC family whose members are conserved in all organisms. Until recently, cohesin was mainly studied for its key role in maintaining sister chromatid cohesion (SCC) after replication, which is necessary for ensuring sister chromatid segregation during chromosome separation.
In addition to this role, numerous studies have recently demonstrated that this complex establishes and maintains chromatin loops in mammals along chromosomes during interphase, i.e. outside cell division periods. It is thought that cohesin captures small loops through its ring, which are then gradually enlarged. These loops appear to be involved in regulating biological processes such as transcription by e.g. enabling distant activating or repressive regions to approach a gene. The mechanisms regulating the formation, extension and positioning of these loops are being actively researched.
Using chromosome conformation capture (Hi-C) on the yeast Saccharomyces cerevisiae, scientists have revealed that this complex is capable of inducing chromatin loops during metaphase (mitosis stage in which replicated chromosomes are aligned before anaphase) rather than interphase. These newly established loops are involved in chromosome compaction, and new protein actors regulating their size have been characterized. For instance, proteins Eco1 and Wpl1, also found in the human cohesin complex, negatively regulate the extension of DNA loops. These are recruited to cohesin by the protein Pds5 through two independent pathways regulating the positioning and size of loops.
This study reveals that, although molecular processes involved in cohesin-regulated loop formation are conserved between humans and yeast, they have been selected over the course of evolution to respond to different biological functions.
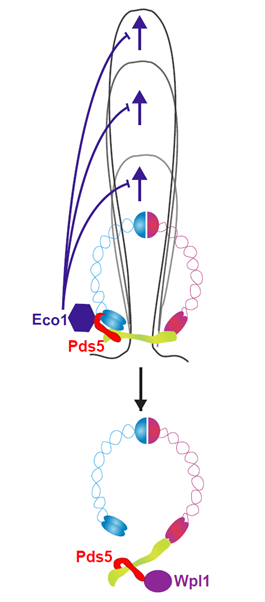
Two activities oppose the expansion of DNA loops: Wpl1 activity dissociates cohesins on DNA, while Eco1 locks the cohesin molecular motor to the advance of cohesins on DNA. These proteins are recruited by Pds5 to the cohesin.
Source : Molecular Cell, March 19 2020 doi: 10.1016/j.molcel.2020.01.019